Abstract
Relapse-related mortality remains one of the main causes of death in patients (pts) with myeloid neoplasms (MN) who undergo allogeneic hematopoietic cell transplantation (allo-HCT). Molecular mutations and cytogenetic abnormalities present in hematopoietic cells are important determinants of relapse risk, with MRD predictive of relapse after allo-HCT. We studied the clonal hierarchy of MN at diagnosis compared to the relapsed state, to determine the effects of the ancestral hit and subclones on the impact on post-transplant recurrence.
DNA from marrow/PB samples of MN pts was subjected to analysis at the time of diagnosis, post, and at the time of relapse to evaluate clonal expansion or evolution using a targeted multi-amplicon deep NGS panel of all ORFs of the top 60 most commonly mutated genes in MDS.
We identified 43 pts with MDS and MDS/MPN who underwent allo-HCT from 2000 to 2016, whom were assessed serially in non-relapsed MN (NR-MN) and relapsed (R-MN). Median age at diagnosis was 54 years (y, range, 22-69) and 54y (range, 22-70) at time of HCT; 56% were male. NR-MN vs. R-MN pts median IPSS and IPSS-R scores at the time of diagnosis, 1.0 vs. 1.25 (P=0.5) and 4.0 vs. 4.0 (P=0.6), respectively. 53% underwent MAC and 58% had a PBSC. Pre-HCT treatments included HMA (42%), ESA (29%), 7+3 (10%), IMiD (10%), and HU (10%). Serial cases had a median follow up of 4 y (range 0.7 - 14.8) from diagnosis (Dx) and 2.3 (0.1-10.7) y from HCT. Rates before HCT of CR (30% vs. 30%), progressive disease (13% vs. 5%), stable disease (25% vs 25%), and disease progression (35% vs. 40%) (P=0.89). Median follow up post allo-HCT was 1.6y vs. 2.5y (P= 0.26). The incidence of acute GVHD in NR-MN vs. R-MN was 43% vs. 65% (P= 0.269) and 39% vs. 15% for chronic GVHD (P= 0.156), respectively. No difference in median blast percentage pre allo-HCT 4% vs 3.5% (P=0.5722).
In NR-MN pts (N=23) after transplant, their pre-HCT characteristics showed 45% (10/23) had abnormal cytogenetics, with the most common abnormalities being del17p, del7q, and/or monosomy 7. SNP analysis showed pathogenic lesions in 43% of assessable cases (6/14), with most frequent abnormalities being loss of 14q11.2 (N=3) and aberrations (loss/UPD) of chr 7 (N=3). Furthermore, 17/21 pts had ≥1 mutation in decreasing frequency TET2 (N=4) , BCOR/BCORL1 (3) , SRSF2 (3), STAG2 (3), ASXL1 (3) , TP53 (2). Post-HCT NGS analyses are ongoing.
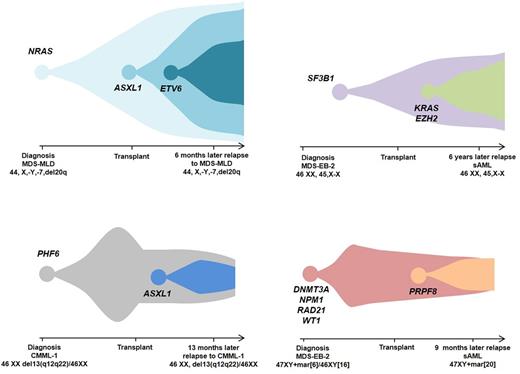
In R-MN pts (n=20) after transplant, their pre-HCT characteristics showed 65% had abnormal cytogenetics at diagnosis, with the most common chromosomal aberrations involving monosomy 7/7q, trisomy 8, del5q, and del20q. Pre-HCT, 14/20 pts had ≥1 mutation in decreasing frequency TP53 (N=4) , SF3B1 (3), U2AF1 (3) , DNMT3A (2) , NRAS (2), and ASXL1 (2) . NGS at disease diagnosis, post-HCT, and at relapse was performed for 9 pts. In 5 pts relapse of the original ancestral hit (PHF6, SF3B1, NRAS, TP53, and DNMT3A) evolution of additional mutations (ASXL1, KRAS, EZH2) (4 featured in Figure 1). Clinical management of relapsed pts included a 2nd allo-HCT in 7 pts (35%), of which 2 are alive, and DLI in 6 (30%), of which 3 are alive.
Logistic regression analysis demonstrated that mutations in DNMT3A and TP53 were non-significantly associated with higher relapse rates (OR= 3.27; P=0.1403). Univariable analysis for time to relapse demonstrated a significant association with earlier relapse with chromosome 11 aberrations (HR=6.4; P=0.005), but DNMT3A and TP53 together (HR=2.285; P=0.1071), TP53 alone (HR=2.257; P=0.1576), and complex cytogenetics (HR=2.15; P=0.1733) with relapse did not reach statistically significance, due to limited sample size.
In conclusion, serial analysis of R-MN pre and post-HCT indicated recurrences of the original ancestral mutation, but often included acquisition of new mutations. Retraction but not elimination of the initial ancestral clone in R-MN pts demonstrated the relevance of an earlier ontogenetic stage. The initial hit could potentially drive the phenotype and resultant characteristics of the disease of subsequent lesions or vice versa. We plan to apply ultra-deep sequencing followed by an in-house bio-analytical pipeline to exclude false positive low frequency hits thereby determining the true MRD for the mutation identified in the original MN pre allo-HCT. Deeper NGS of MN pts in the pre & post allo-HCT setting will expand understanding of primary clonal aberrations and their subsequent re-emergence or regression with disease evolution.
Gerds: CTI BioPharma: Consultancy; Incyte: Consultancy. Advani: Pfizer: Consultancy; Takeda/ Millenium: Research Funding. Sekeres: Celgene: Membership on an entity's Board of Directors or advisory committees. Majhail: Sanofi: Honoraria; Anthem, Inc.: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal